Transcriptomics and Immune Repertoire (TCR & BCR)
Relying on 15 years of research, our regular immune repertoire analysis at Parean are not the common ones available.
After QC and preprocessing, we provide descriptive gene and diversity profiles, spectra typing analysis or repertoire sharing analysis

Deliverables
Tables including count data in MiAIRR format.
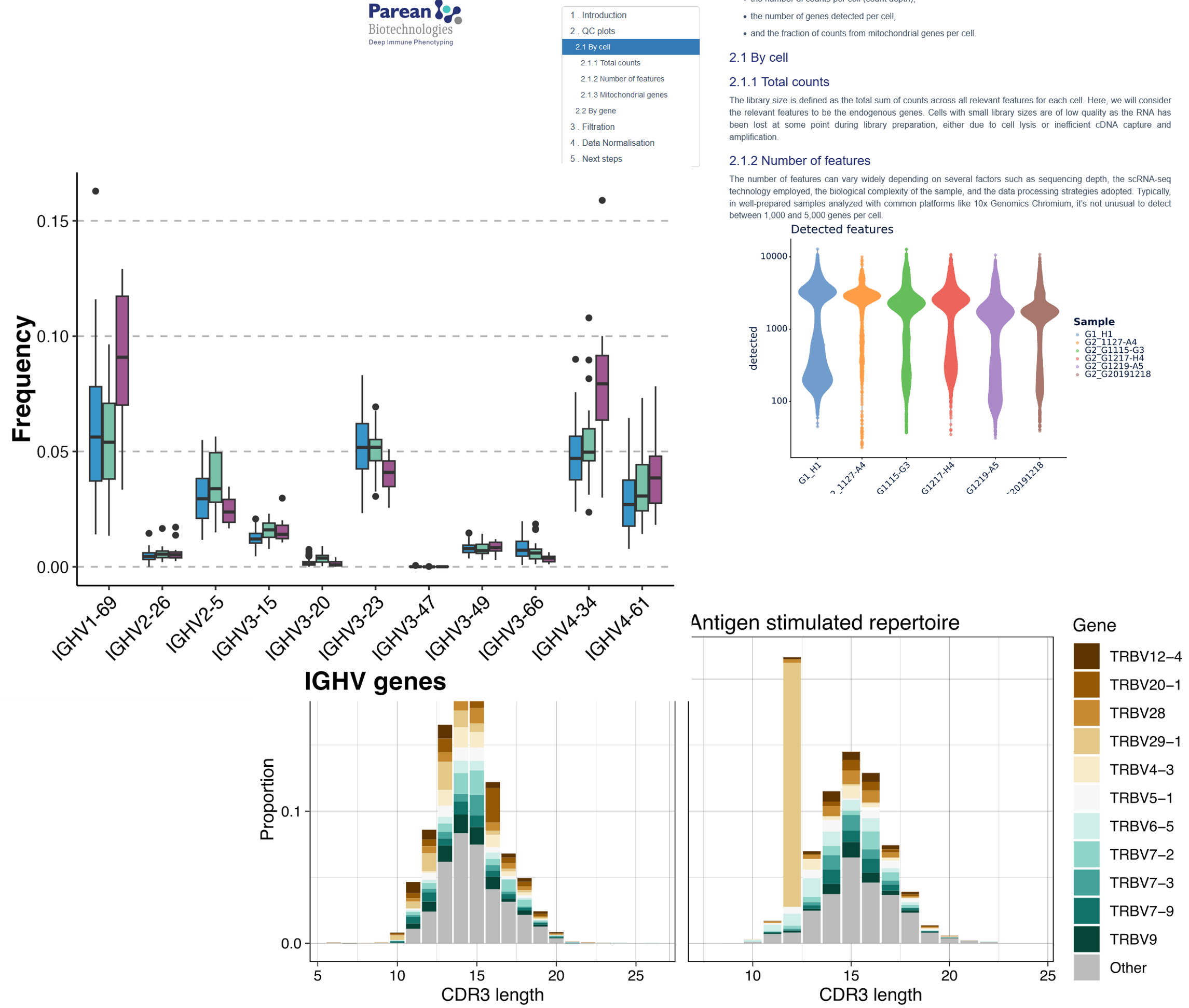
Descriptive gene usage (V, J & V-J),
Descriptive diversity profiles (Richness, Shannon entropy, Mean clonal frequency),
Spectratyping analysis (CDR3 length distributions),
Repertoire sharing analysis (clonal overlap).
Deliverables
Tables including count data in MiAIRR format & Graphs, meeting with our team.
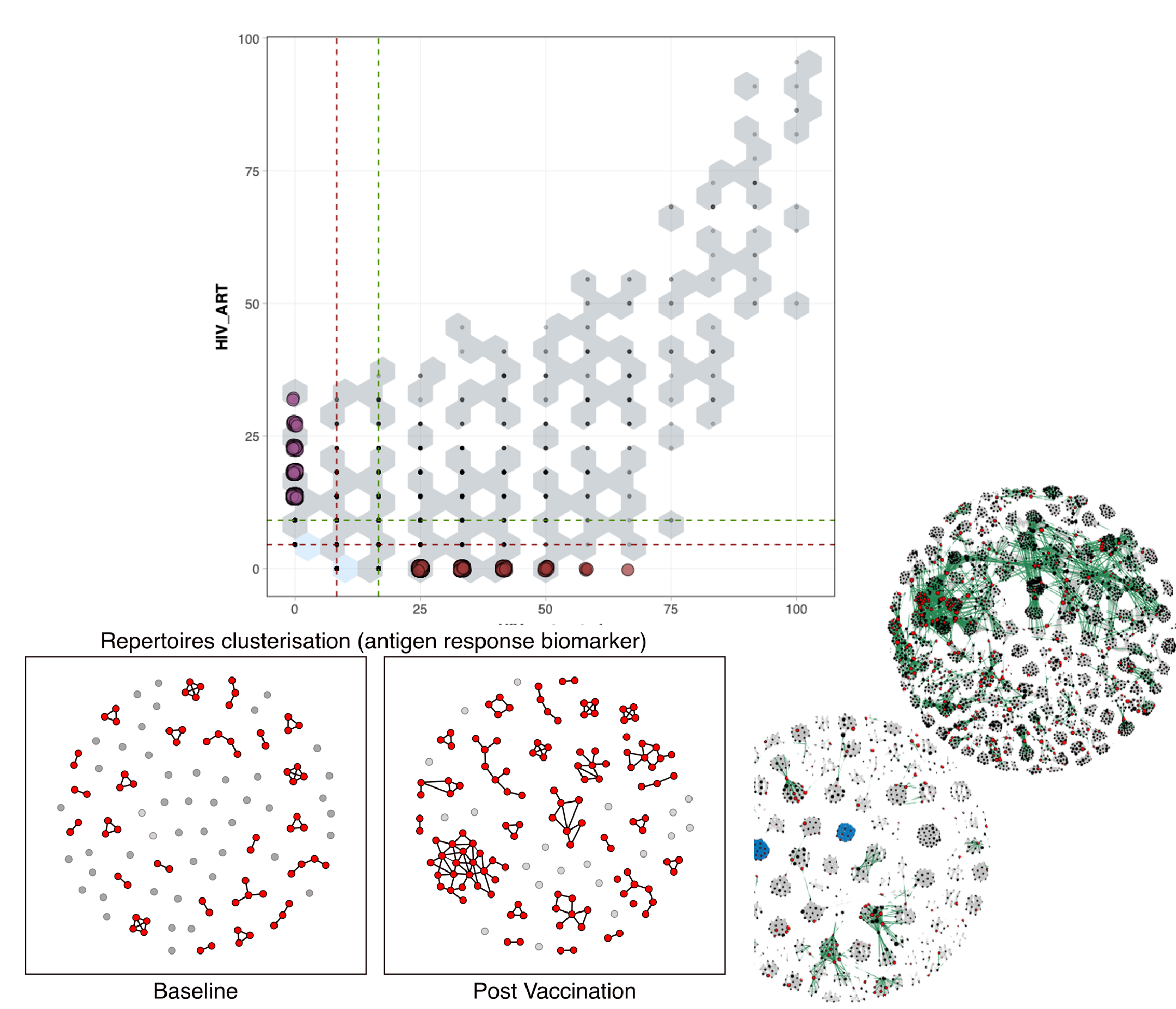
We master cutting-edge immune repertoire analysis, powered by AI.
Dive into the best analysis and representation such diversity metrics, repertoire sharing analysis, clonal architecture, generation probability and clonal specificity, in silico repertoire simulation or public datasets checking.

Deliverables
Bioinformatics object, complemented by the analyses listed above and relevant graphs
Your time is precious, and so are your samples. Our regular transcriptomics protocols enables robust and fast analysis of gene expression, trimming if needed, exploratory analysis and relevant graphical representation.

Raw Data QC, Trimming if needed, Alignment on latest reference genome, Quantification of sequences into count data.
Deliverables
Fastq; QC report; Raw count data and meeting with our team.
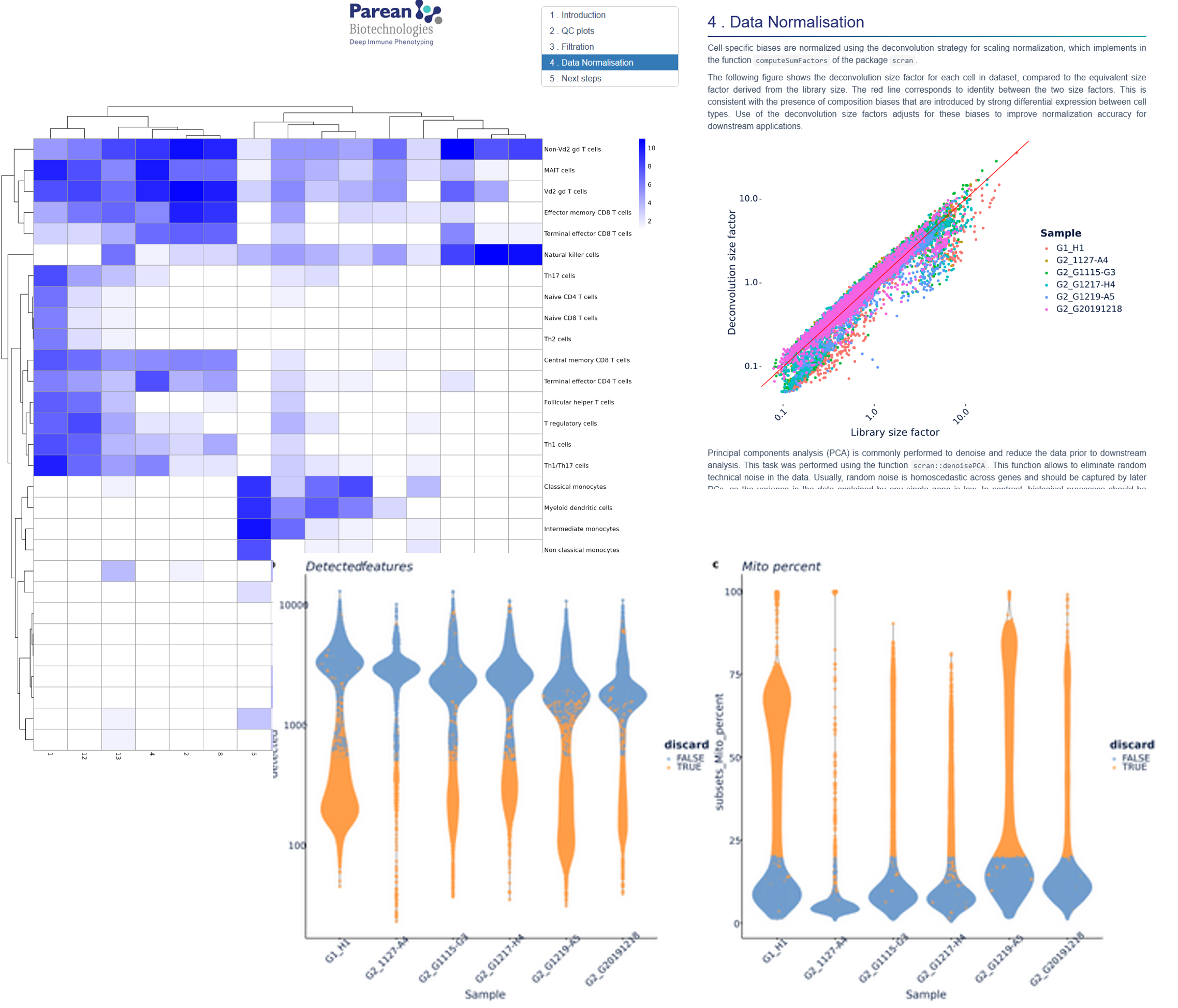
Data transformation and normalisation, Exploratory Analysis & Filtration.
Deliverables
Normalized data; Integrated sample data; Exploratory analysis figures (+Fastq; QC report; Raw count data); meeting with our team.
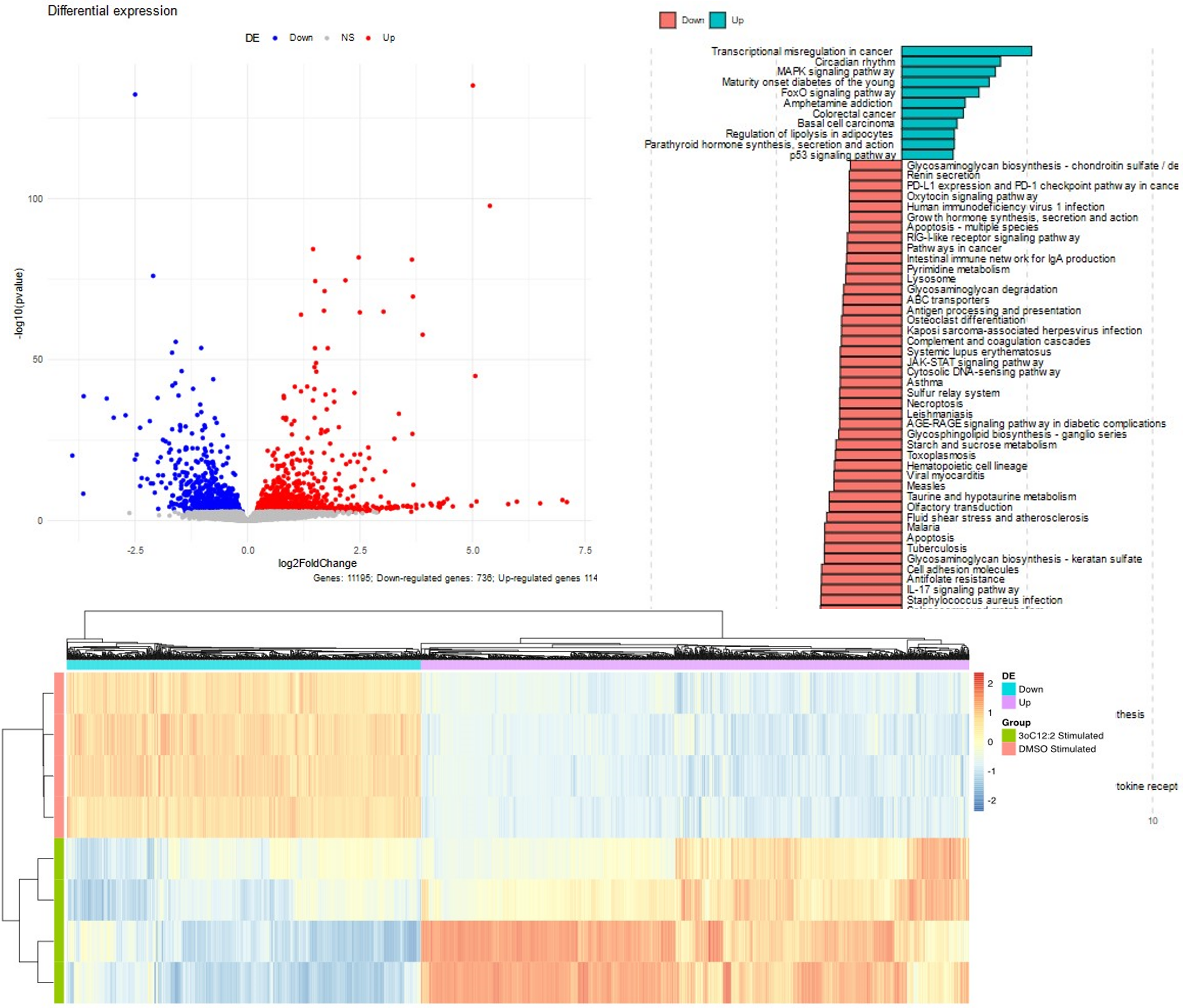
Go beyond regular analysis, and use our methods for batch effect correction, and use relevant methods such as Over Representation Analysis, Gee Set analysis (public or specific signature), differential gene expression, HLA typing…

Deliverables
Graphical representation; meeting with our team (+ standard analysis deliverables).

Integrated technological platform, dedicated to cell phenotyping, omics generation and data analysis.
Go to Wet Labs